ZOOHCC - 501: Molecular Biology (Theory)
Unit 3: Transcription and Regulatory RNAs
Unit 3: Transcription and Regulatory RNAs
Transcription in Eukaryotes
Prokaryotes and eukaryotes perform basically the same transcription process, but there are some important differences. The most important difference between prokaryotes and eukaryotes is the membrane-bound nucleus and organelles of the latter. Eukaryotic cells, which have genes encapsulated in the nucleus, must be able to transport mRNA to the cytoplasm and must protect the mRNA from degradation before it is translated. Eukaryotes also use three different polymerases, each of which transcribes a different subset of genes.
Initiation
The eukaryotic promoters that we are most interested in are similar to prokaryotic promoters in that they contain a TATA box (Figure 1). However, transcription initiation in eukaryotes is much more complex than in prokaryotes. Unlike prokaryotic RNA polymerase, which can itself bind to a DNA template, eukaryotes require several other proteins, called transcription factors, to first bind to the promoter region and then recruit the appropriate polymerase. help you to
In addition, there are three different RNA polymerases in eukaryotes, each composed of ten or more subunits. Each eukaryotic RNA polymerase also requires a specific set of transcription factors to reach the DNA template.
RNA polymerase I
RNA polymerase I resides in the nucleolus, a specialized nuclear substructure where ribosomal RNA (rRNA) is transcribed, processed, and assembled into ribosomes. rRNA molecules are considered structural RNAs because they perform cellular functions but are not translated into proteins. rRNA is a component of the ribosome and is essential for the translation process. RNA polymerase I synthesizes most rRNA.
RNA polymerase II
RNA polymerase II resides in the cell nucleus and synthesizes all nuclear pre-mRNAs that code for proteins. Eukaryotic pre-mRNAs undergo extensive post-transcriptional and pre-translational processing. For clarity, the term 'mRNA' is only used to denote the mature, processed molecule that is ready for translation. RNA polymerase II is involved in the transcription of most eukaryotic genes.
RNA polymerase III
RNA polymerase III is also located in the cell nucleus. This polymerase transcribes a variety of structural RNAs, including transfer pre-RNAs (pre-tRNAs) and small core pre-RNAs. tRNAs play an important role in translation. They act as adapter molecules between the mRNA template and the growing polypeptide chain. Small nuclear RNAs have a variety of functions, including 'splicing' pre-mRNAs and regulating transcription factors.
Each type of RNA polymerase recognizes different promoter sequences and requires different transcription factors.
Elongation
After formation of the pre-initiation complex, the polymerase is released from other transcription factors, elongation proceeds in a manner similar to prokaryotes, and RNA polymerase synthesizes mRNA in the 5' to 3' direction. As mentioned earlier, RNA polymerase II transcribes the majority of eukaryotic genes, so this section will focus on how this polymerase accomplishes elongation and termination.
Although the enzymatic elongation process is essentially the same in eukaryotes and prokaryotes, the DNA template is more complex. When eukaryotic cells are not dividing, their genes exist as a diffuse mass of DNA and protein called chromatin. DNA clusters around charged histone proteins at repeated intervals. Collectively called nucleosomes, these DNA-histone complexes are regularly spaced and consist of 146 DNA nucleotides wrapped around eight histones like threads around a spool.
For RNA synthesis to take place, histones must be eliminated each time the transcription machinery encounters a nucleosome. This is achieved by a special protein complex called FACT. It stands for “promoting chromatin transcription.” This complex pulls the histone away from the DNA template as the polymerase moves along it. Once the pre-mRNA is synthesized, the FACT complex replaces histones to reassemble the nucleosome.
Termination
Termination of transcription depends on the polymerase. In eukaryotes, unlike in prokaryotes, extension by RNA polymerase II occurs 1,000–2,000 nucleotides after the end of the transcribed gene. This pre-mRNA tail is removed during mRNA processing. RNA polymerases I and III require termination signals. Genes transcribed by RNA polymerase I contain a specific 18-nucleotide sequence that is recognized by a termination protein. The termination process of RNA polymerase III involves an mRNA hairpin that triggers the release of the mRNA
Eukaryotic RNA Polymerases and General Transcription Factors
Although the basic mechanisms of transcription are the same in all cells, they are considerably more complex in eukaryotic cells than in bacteria. This is reflected in her two distinct differences between prokaryotic and eukaryotic systems. First, all genes in bacteria are transcribed by a single RNA polymerase, whereas eukaryotic cells contain several different RNA polymerases that transcribe different classes of genes. Second, eukaryotic RNA polymerases must specifically initiate transcription by interacting with various additional proteins rather than directly binding to promoter sequences. This increased complexity of eukaryotic transcription likely facilitates the sophisticated regulation of gene expression necessary to direct the activities of many different cell types in multicellular organisms..
Protein involvement in prokaryotic transcription
Eukaryotic cells contain three different nuclear RNA polymerases that transcribe different classes of genes (Table 6.1). Protein-coding genes are transcribed by RNA polymerase II to produce mRNA. Ribosomal RNA (rRNA) and transfer RNA (tRNA) are transcribed by RNA polymerases I and III. RNA polymerase I specializes in transcription of the three largest types of rRNA, named 28S, 18S, and 5.8S, depending on their sedimentation velocities during high-speed centrifugation. RNA polymerase III transcribes genes to tRNA and minimal ribosomal RNA (5S rRNA). Some of the small RNAs (snRNAs and scRNAs) involved in splicing and protein transport are also transcribed by RNA polymerase III, while others are polymerase II transcripts. In addition, another RNA polymerase (similar to that of bacteria) is found in chloroplasts and mitochondria and specifically transcribes the DNA of these organelles.
All three nuclear RNA polymerases are complex enzymes, each composed of 8–14 different subunits. They recognize different promoters and transcribe different classes of genes, but share some common features. The two largest subunits of all three eukaryotic RNA polymerases are related to the single E. coli RNA polymerase β and β' subunits. Furthermore, all three different enzymes share five subunits of eukaryotic RNA polymerases. Consistent with these structural similarities, the various eukaryotic polymerases share several functional properties, such as the need to interact with other proteins to properly initiate transcription.
General Transcription Factors and Initiation of Transcription by RNA Polymerase II
RNA polymerase II is the focus of most transcriptional studies in eukaryotes because it is responsible for synthesizing mRNA from protein-coding genes. Early attempts to study this enzyme showed that its activity differs from that of prokaryotic RNA polymerases. Accurate transcription of bacterial genes can be achieved in vitro by simply adding purified RNA polymerase to promoter-containing DNA, but is not possible in eukaryotic systems. The basis for this difference was elucidated in 1979 when Robert Roeder and his colleagues discovered that RNA polymerase II could initiate transcription only when additional proteins were added to the reaction. Thus, transcription in eukaryotic systems appeared to require various initiation factors that are not associated with polymerases (in contrast to bacterial σ factors).
Biochemical fractionation of nuclear extracts has identified specific proteins (called transcription factors) required for RNA polymerase II to initiate transcription. Indeed, the identification and characterization of these factors represent a major part of ongoing efforts to understand transcription in eukaryotic cells. Two general types of transcription factors have been defined. General transcription factors form part of the basic transcription machinery as they are involved in transcription from all polymerase II promoters. Additional transcription factors (described later in this chapter) are involved in the regulation of gene expression as they bind to DNA sequences that control the expression of individual genes.
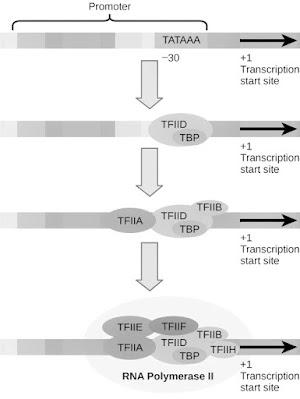
Initiation of transcription by RNA polymerase II in the reconstituted in vitro system requires five general transcription factors (Figure 6.12). The promoters of many genes transcribed by polymerase II contain a TATAA-like sequence 25-30 nucleotides upstream of the transcription start site. This sequence (termed the TATA box) resembles the -10 sequence element of bacterial promoters, and mutations in the TATAA sequence demonstrated their role in transcription initiation. The first step in transcription complex formation is the binding of a common transcription factor called TFIID to his TATA box (TF stands for transcription factor and II for polymerase II). TFIID itself is composed of multiple subunits, including the TATA-binding protein (TBP), which specifically binds to the TATAA consensus sequence, and 10–12 other polypeptides called TBP-associated factors (TAFs). TBP then binds to a second common transcription factor (TFIIB) to form a TBP-TFIIB complex at the promoter (Figure 6.13). TFIIB then acts as a bridge to RNA polymerase, which, along with her third factor, TFIIF, binds to her TBP-TFIIB complex.
After recruitment of RNA polymerase II to the promoter, transcription initiation requires the binding of two additional factors (TFIIE and TFIIH). TFIIH is a multi-subunit factor that appears to play at least two important roles. First, her two subunits of TFIIH are helicases and can unwrap DNA around the initiation site. (These subunits of TFIIH are also required for nucleotide excision repair, as discussed in Chapter 5.) Another subunit of TFIIH is the repeat located in the C-terminal domain of the largest subunit of RNA polymerase II. A protein kinase that phosphorylates sequences. Phosphorylation of these sequences is thought to disassociate the polymerase from the initiation complex and allow it to proceed down the template, elongating the growing RNA strand.
In addition to the TATA box, the promoters of many genes transcribed by RNA polymerase II contain a second critical sequence element (initiator or Inr sequence) spanning the transcription start site. In addition, some RNA polymerase II promoters contain only Inr elements without TATA boxes. Initiation at these promoters still requires TFIID (and TBP), but TBP does not appear to recognize these promoters by binding directly to her TATA sequences. Instead, other TFIID subunits (TAFs) appear to bind to her Inr sequences. This binding recruits TBP to the promoter and assembles TFIIB, polymerase II, and additional transcription factors as previously described. Thus, TBP plays a central role in the initiation of polymerase II transcription, even on promoters lacking a TATA box.
Despite the development of in vitro systems and the characterization of several general transcription factors, much remains unknown about the mechanisms of polymerase II transcription in eukaryotic cells. The sequential recruitment of transcription factors described here represents a minimal system required for transcription in vitro. Additional factors may be required within the cell. Furthermore, RNA polymerase II appears to be able to associate with several transcription factors in vivo before the transcription complex is assembled on DNA. In particular, preformed complexes of RNA polymerase II with TFIIB, TFIIE, TFIIF, TFIIH and other transcriptional regulatory proteins have been detected in both yeast and mammalian cells. These large complexes, called polymerase II holoenzymes, are recruited to promoters by direct interaction with TFIID (Figure 6.14). Therefore, the relative contribution of the stepwise assembly of individual factors to the recruitment of the RNA polymerase II holoenzyme to promoters in cells remains to be determined.
Transcription by RNA Polymerases I and III
As previously mentioned, various RNA polymerases are involved in the transcription of genes encoding eukaryotic ribosomes and transfer RNAs. However, all three RNA polymerases require additional transcription factors to associate with appropriate promoter sequences. Furthermore, although the three different polymerases recognize different types of promoters in eukaryotic cells, a common transcription factor, the TATA-binding protein (TBP), appears to be required for transcription initiation by all three enzymes. .
RNA polymerase I is dedicated to transcription of ribosomal RNA genes present in tandem repeats. Transcription of these genes produces a large 45S pre-rRNA, which is processed to produce 28S, 18S, and 5.8S rRNA (Figure 6.15). The promoter of the ribosomal RNA gene extends approximately 150 base pairs immediately upstream of the transcription start site. These promoter sequences are recognized by two transcription factors, UBF (upstream binding factor) and SL1 (selectivity factor 1), which cooperatively bind to the promoter and recruit polymerase I to form the initiation complex. (Figure 6.16). The SL1 transcription factor is composed of four protein subunits, one of which surprisingly is TBP. The role of TBP was directly demonstrated by the finding that yeast harboring mutations in TBP are defective not only in transcription by polymerase II, but also in transcription by polymerases I and III. Thus, TBP is a common transcription factor required for all three classes of eukaryotic RNA polymerases. Because the promoters of ribosomal RNA genes do not contain TATA boxes, TBP does not bind to specific promoter sequences. Instead, the binding of TBP to ribosomal RNA genes is mediated by the binding of other proteins of the SL1 complex to their promoters. This is a similar situation to the binding of TBP with the Inr sequence of the polymerase II gene, where the TATA box is missed.
tRNA, 5S rRNA, and several small RNA genes involved in splicing and protein transport are transcribed by polymerase III. These genes are characterized by promoters located internal to the transcribed sequence rather than upstream. The most thoroughly studied gene transcribed by polymerase III is his 5S rRNA gene in Xenopus laevis. TFIIIA, the primary purified transcription factor, initiates assembly of the transcription complex by binding to specific DNA sequences in the 5S rRNA promoter. This binding is followed by the sequential binding of TFIIIC, TFIIIB, and polymerase. The tRNA gene promoter differs from the 5S rRNA promoter in that it does not contain the DNA sequence recognized by TFIIIA. Instead, TFIIIC binds directly to the promoter of the tRNA gene and recruits TFIIIB and polymerase to form a transcription complex. TFIIIB is composed of several subunits, of which (again) the TATA-binding protein is TBP. Although her three RNA polymerases in eukaryotic cells recognize different promoters, TBP appears to be the common element linking promoter recognition to polymerase recruitment to transcription complexes.